Design of New Bioinspired Superantibiotics
 The introduction of
antibiotics as a means to treat and control infectious diseases marked a
turning point in the evolution of modern medicine. However, misuse of
these antimicrobial agents has caused the development of widespread
antibiotic resistance (ABR) in organisms ranging from spoilage
microorganisms to pathogens. ABR represents a global public health
threat that must be faced, otherwise it will have disastrous
consequences for global public health. ABR bacterial infections killed
1.27 million people in the world in 2019, Africa being the most affected
continent. Thus, if this growth continues, it is expected by 2050
ABR-related deaths will exceed those due to cancer.
The introduction of
antibiotics as a means to treat and control infectious diseases marked a
turning point in the evolution of modern medicine. However, misuse of
these antimicrobial agents has caused the development of widespread
antibiotic resistance (ABR) in organisms ranging from spoilage
microorganisms to pathogens. ABR represents a global public health
threat that must be faced, otherwise it will have disastrous
consequences for global public health. ABR bacterial infections killed
1.27 million people in the world in 2019, Africa being the most affected
continent. Thus, if this growth continues, it is expected by 2050
ABR-related deaths will exceed those due to cancer.
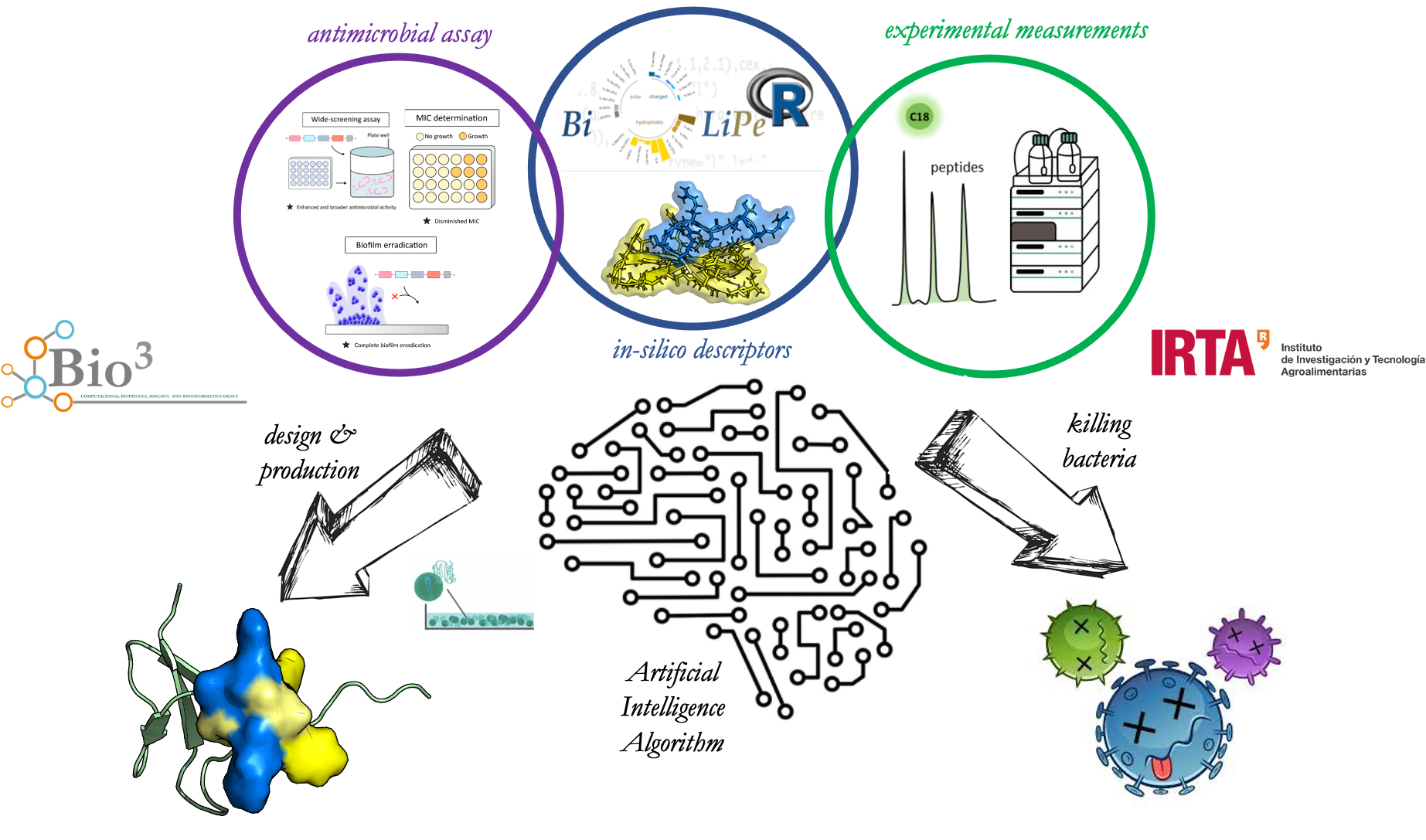
In collaboration with the IRTA - Institute of Agrifood Research and Technology (Dra. Elena García and Dra. Ana Aris), we are design new bioinspired superantibiotics based on personalized host defense peptides (HDPs) that have higher potency, selectivity, and less toxicity than existing ones to prevent and treat infectious diseases caused by multiresistance or extensive drug resistance (MDR/XDR). Our research project aims recombinantly produce the best-tailored HDPs-based superantibiotics selected by an artificial intelligence algorithm based on a transdisciplinary approach that includes a broad screening antimicrobial assay and experimental and in-silico physicochemical characterization of the HDPs.
Simulation
Drug Design 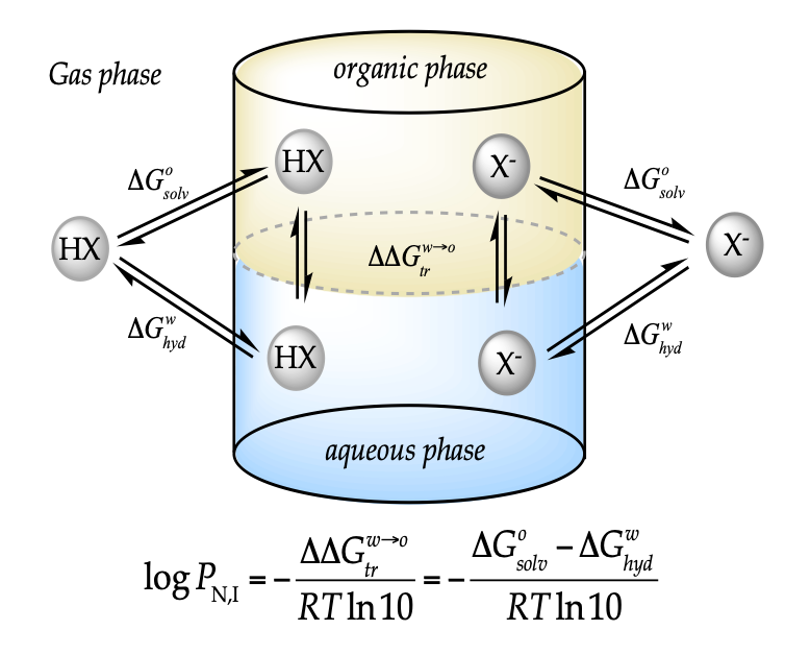 Implementation
of tools for collaborations in the area of medicinal chemistry in
antibiotic development such as use and development of chemoinformatics
tools, ADME-Tox properties, docking using Glide, QSAR studies and
classical molecular dynamic simulations using AMBER).
Implementation
of tools for collaborations in the area of medicinal chemistry in
antibiotic development such as use and development of chemoinformatics
tools, ADME-Tox properties, docking using Glide, QSAR studies and
classical molecular dynamic simulations using AMBER).
Computational Biology  QSAR studies
and classical molecular dynamic simulations in peptides and proteins,
mainly AMPs. Prediction of the effect of mutations in diseases such as
Alzheimer and Myotonia. Study of peptides with neuroprotective and
antioxidant effects.
QSAR studies
and classical molecular dynamic simulations in peptides and proteins,
mainly AMPs. Prediction of the effect of mutations in diseases such as
Alzheimer and Myotonia. Study of peptides with neuroprotective and
antioxidant effects.
Computational Chemistry 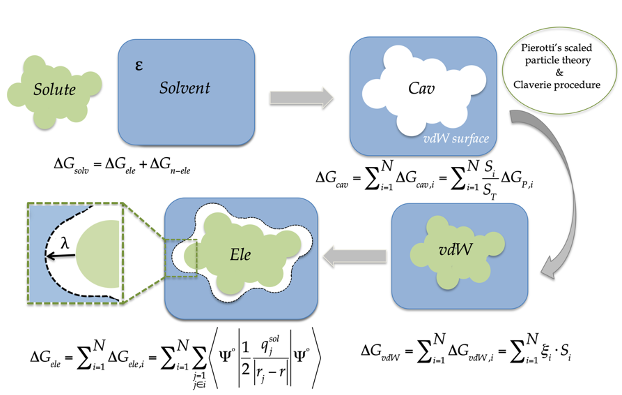 Development and parameterization of the continuous solvation model
IEFPCM/MST using Gaussian. Determination of QM-based physicochemical
properties in small molecules including drugs and pollutants
Development and parameterization of the continuous solvation model
IEFPCM/MST using Gaussian. Determination of QM-based physicochemical
properties in small molecules including drugs and pollutants
Theoretical
Physical chemistry 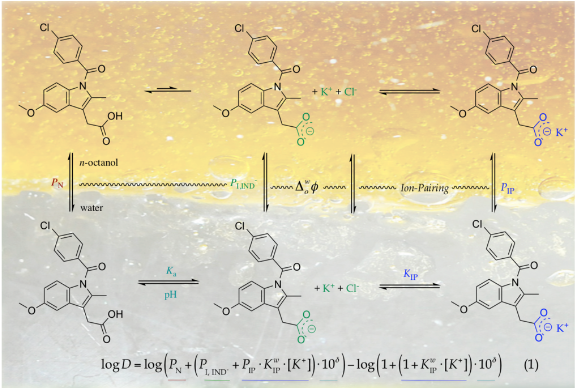 Theoretical
models for lipophilicity profiles of ionizable compounds considering the
effect of the pH and background salt.
Theoretical
models for lipophilicity profiles of ionizable compounds considering the
effect of the pH and background salt.  Models
to explain the relative water content in aerosols as a function of the
organic solute present in these colloidal suspensions.
Models
to explain the relative water content in aerosols as a function of the
organic solute present in these colloidal suspensions.
Drug Design 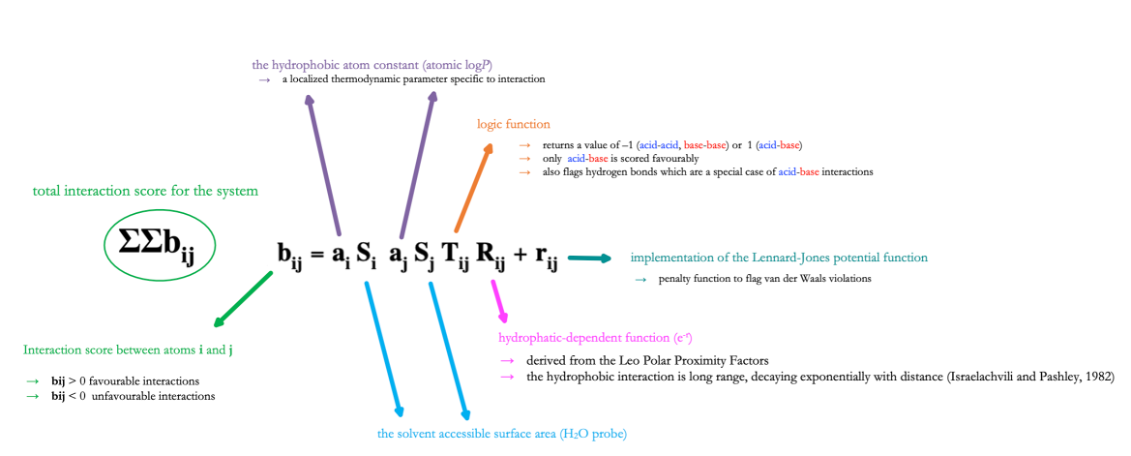 Development of
software and scoring functions for docking studies in collaboration with
Phamacelera Inc.
Development of
software and scoring functions for docking studies in collaboration with
Phamacelera Inc.
Computational Biology 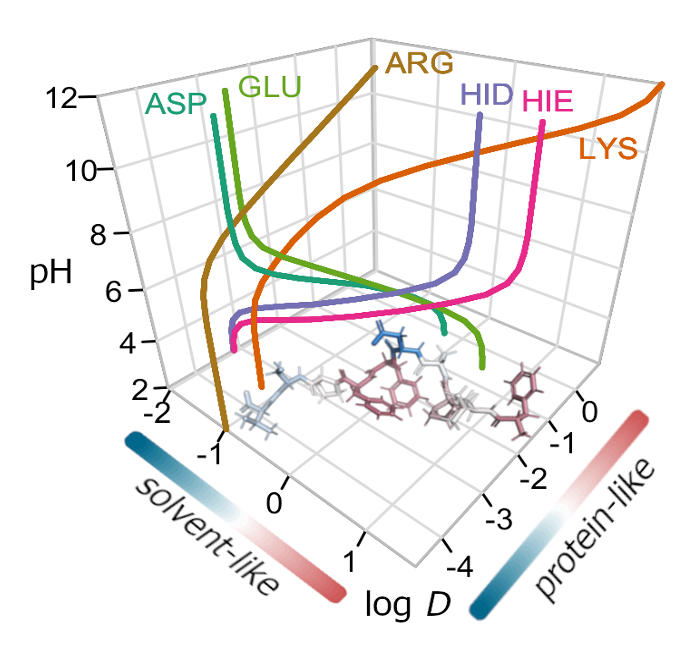 Developed a
structure-based lipophilicity scale of amino acids, models to describe
amphipathicity in antimicrobial peptides-AMPs.
Developed a
structure-based lipophilicity scale of amino acids, models to describe
amphipathicity in antimicrobial peptides-AMPs.
Data Exploration
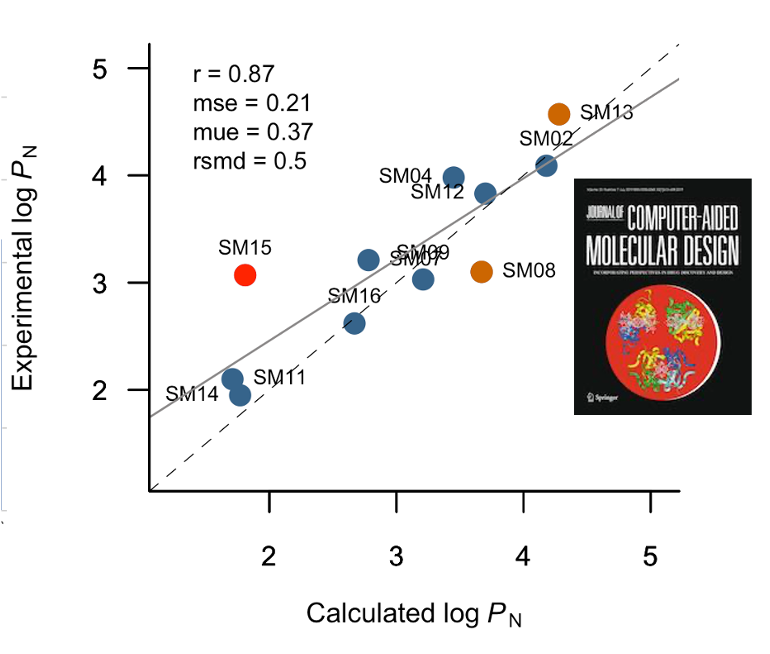 Development of models based on artificial
intelligence for determining physicochemical properties in small
molecules including drugs (bioactivity, lipophilicity and tautomeric
equilibria) and pollutants (solubility and toxicity).
Development of models based on artificial
intelligence for determining physicochemical properties in small
molecules including drugs (bioactivity, lipophilicity and tautomeric
equilibria) and pollutants (solubility and toxicity).
Experimental
 Drug Design
Drug Design
Potentiometric and Shake-flask methods to determine the pKa and lipophilicity of small molecules.
Environmental Pollutants
Quantification by analytical techniques of emerging pollutants for the creation of databases for computational models.
Phytotherapeutics
Analysis, biological activity, collaborative research in the improvement of bioavailability with the application of nanotechnology. Natural Products Research (Structural elucidation methods as NMR, MS, IR)
Separation methods
Column and thin layer chromatography, UPLC, HPLC; Antioxidant methods: Oxygen radical absorption capacity based on fluorometry -ORAC.